version 2.0
Home | Browse | Gene Ontology | Team | Disclaimer | Help
Welcome to SNPRBb (SNP resources for Bubalus bubalis)
SNPRBb Database is Trait specific SNP resources of Bubalus bubalis. The different number of samples from the four traits of the Bubalus bubalis is chosen for finding the SNP which are as following:
| S.No. |
Trait |
Total No. of Samples |
|---|---|---|
| 1. |
Milk Volume |
25 |
| 2. | Age at First Calving |
19 |
| 3. | Post Partum Cyclicity |
19 |
| 4. | Feed Conversion Efficiency |
22 |
Viewing the data
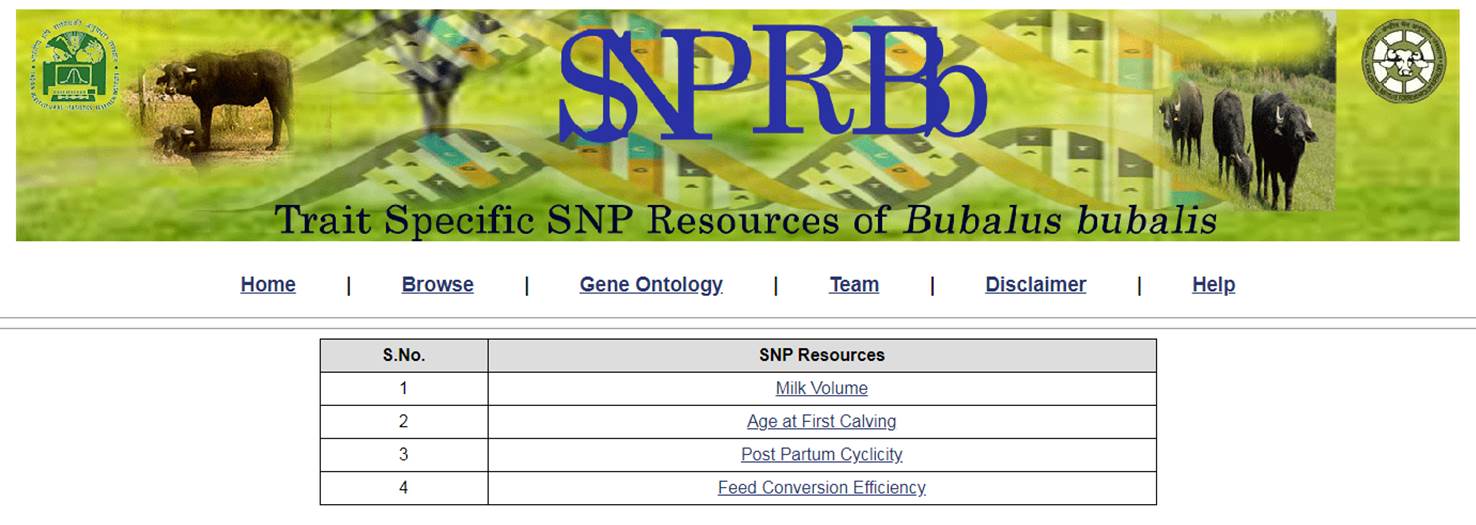
1. The home page of the SNPRBb looks like as follows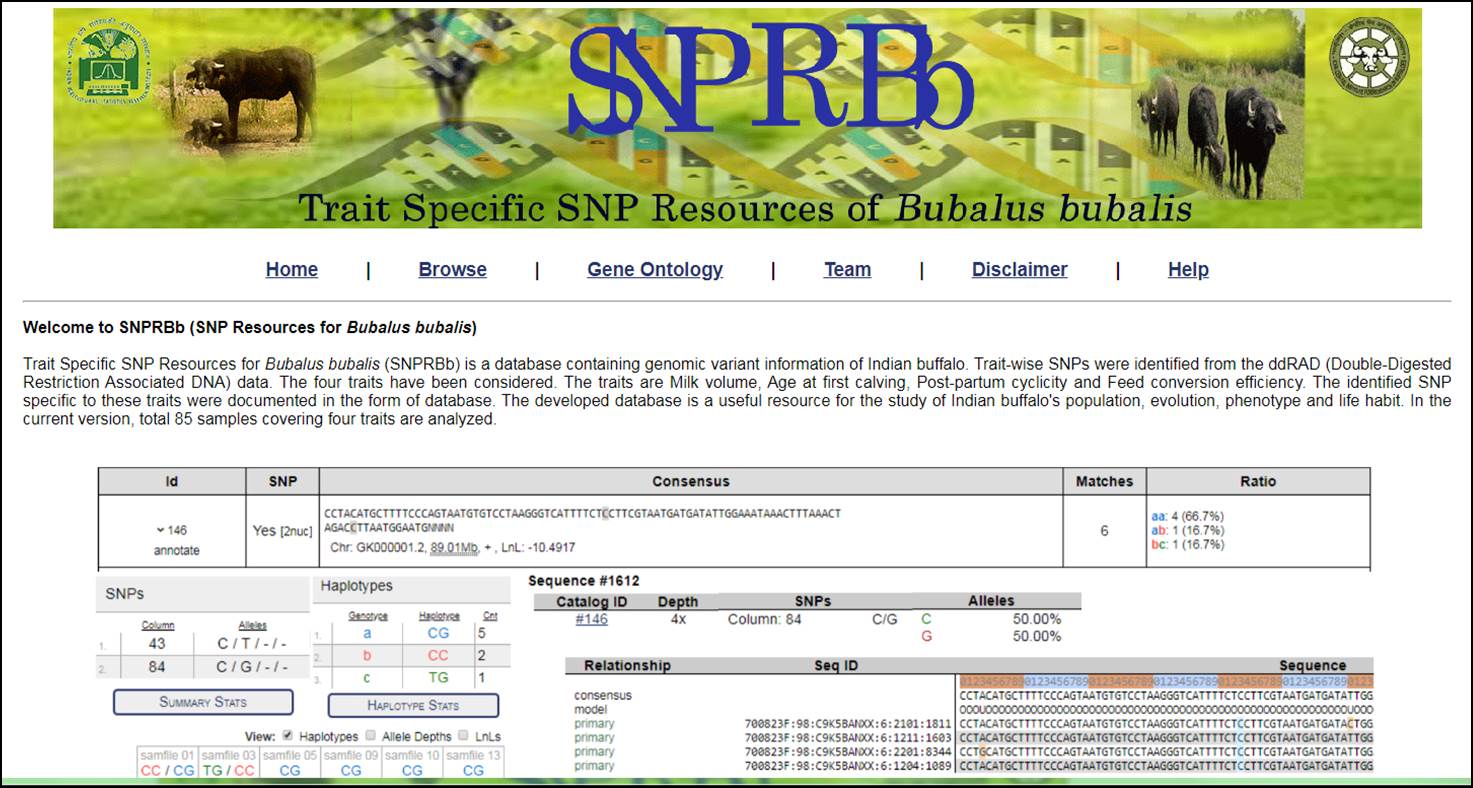
2. The user can click on any of the link i.e., Browse, Team, Disclaimer and Help to see the desired information. If the user wanted to come back to home page at any point of time, he can click on the Home link. The identified SNP resources can be viewed or explored by clicking on the link Browse. This form allows the user to select search the resource based on Catalog and Samples for a given trait. The current database contains the identified SNP resources for four important traits of Bubalus bubalis.
3. Now the user have two options to browse the SNP resources either by choosing the Catalog or Samples for the specific trait. Catalog will provide overall identified Tags, SNPs, Consensus sequence based on Samples (Chromosome Number, Position of Base Pair and LnL), Matches and Allele Ratio in the selected trait. The user can further click on particular Id to get the information related to SNP position, Haplotypes, Genotypes and their count, Allele Depth, LnLs (Log Likelihood filter). In case of more than one SNP, the Summary Statistics can also be viewd by clicking on the SUMMARY STATISTICS. This will display Population, Base Pair, Column, Allele, Alternate Allele, and many other useful information related to the identified SNPs. Similarly, the information is also accessible for HAPLOTYPES STATS in separate window. The step-wise procedure for selection of mentioned parameters and statistics are described below.
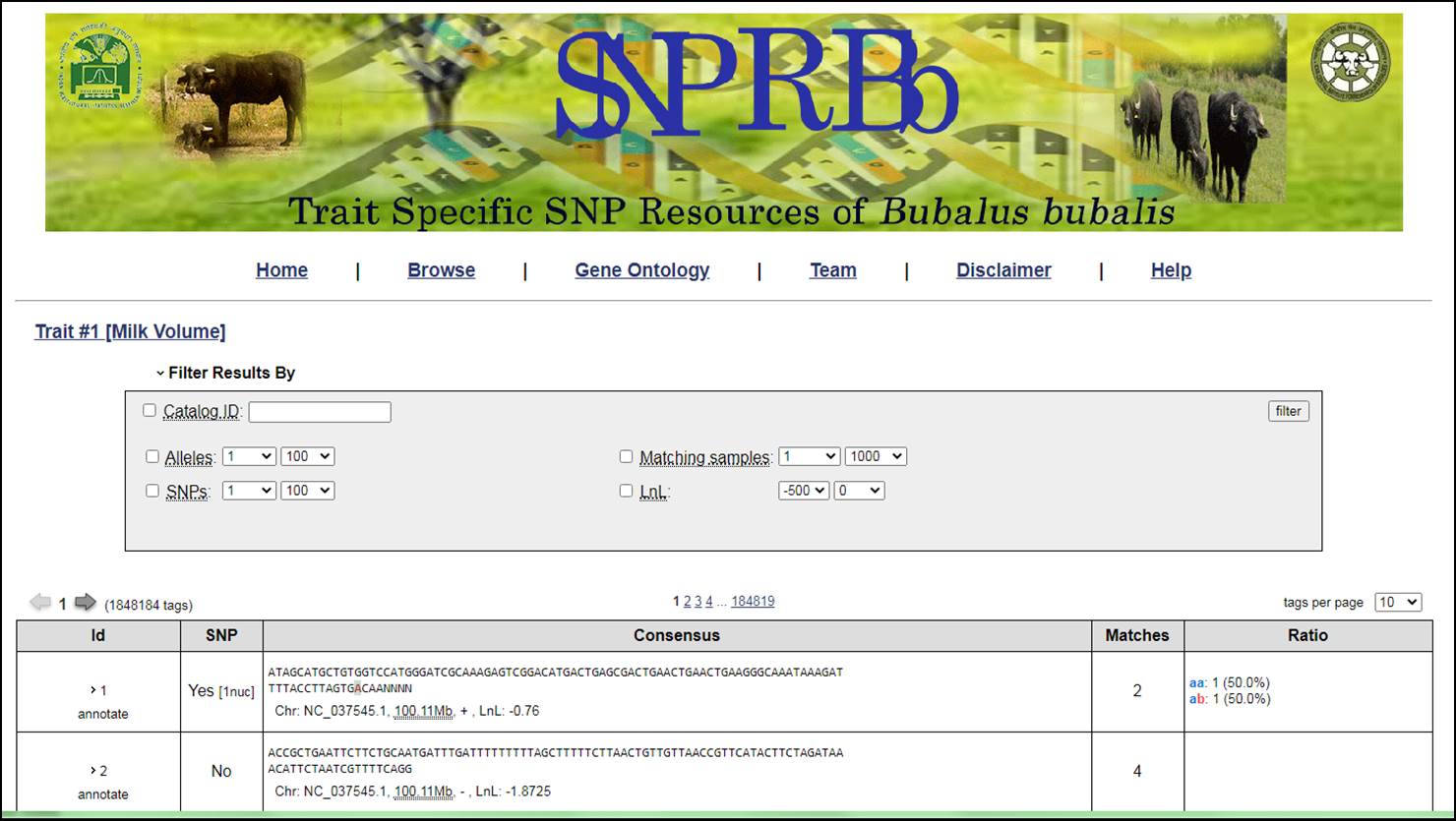
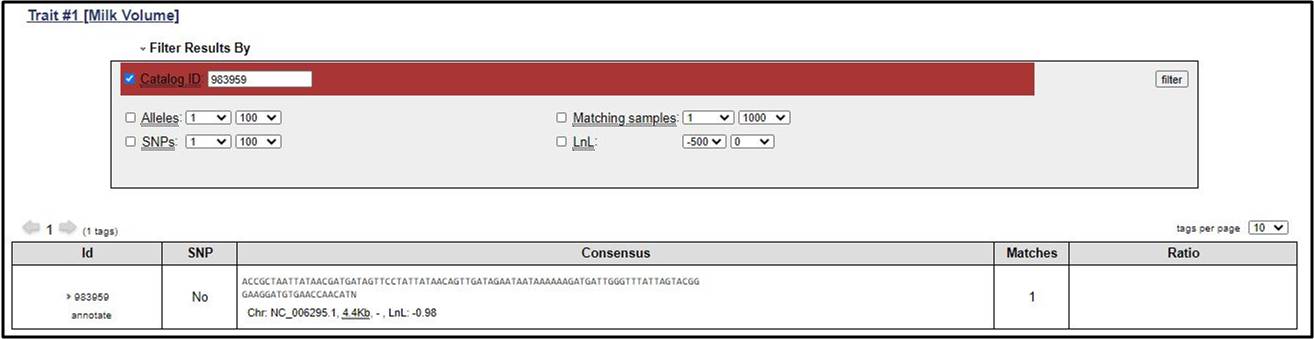
4. The SNP can also be filtered out based on some criteria defined by users. There is need to referesh the results after applying any filter.
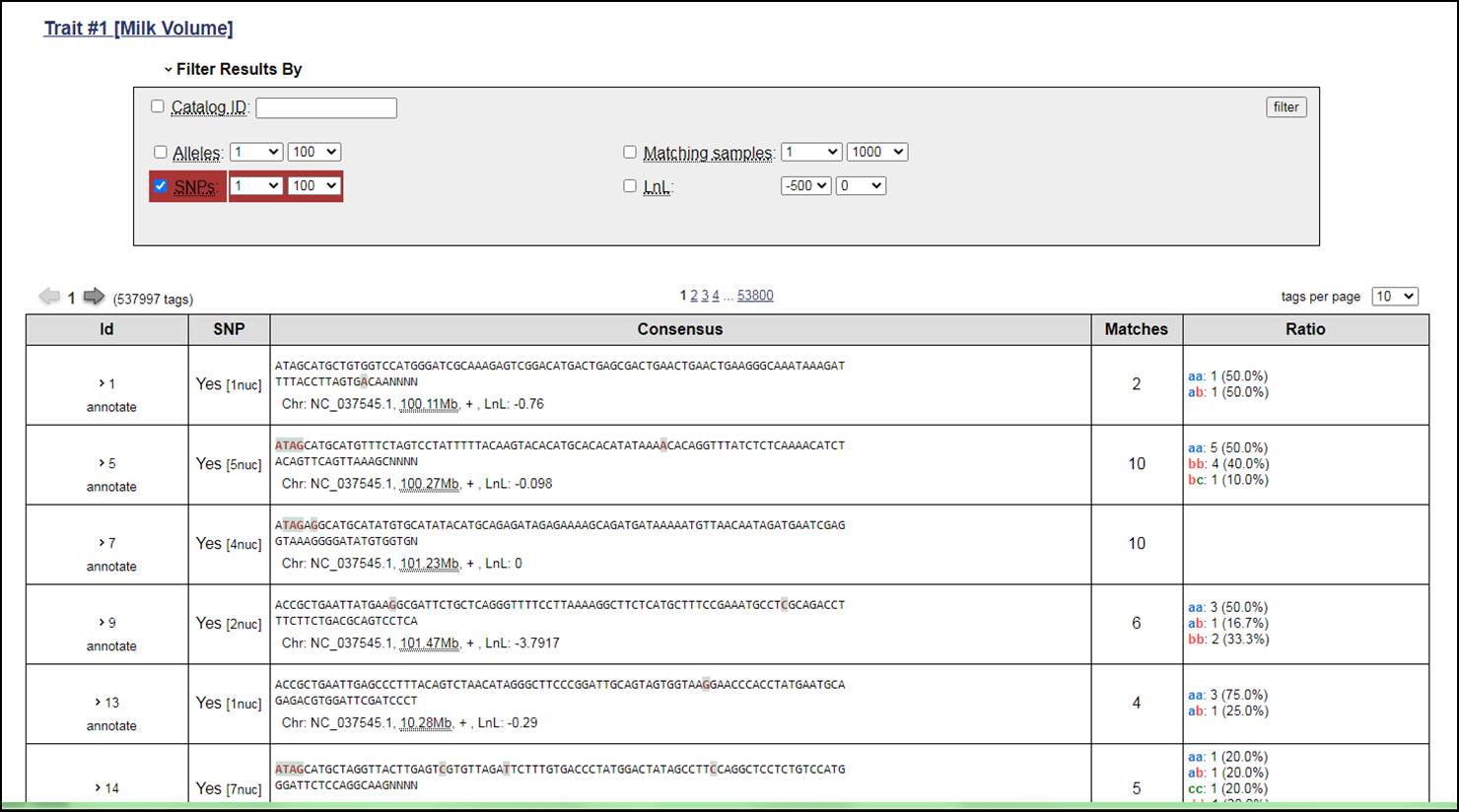
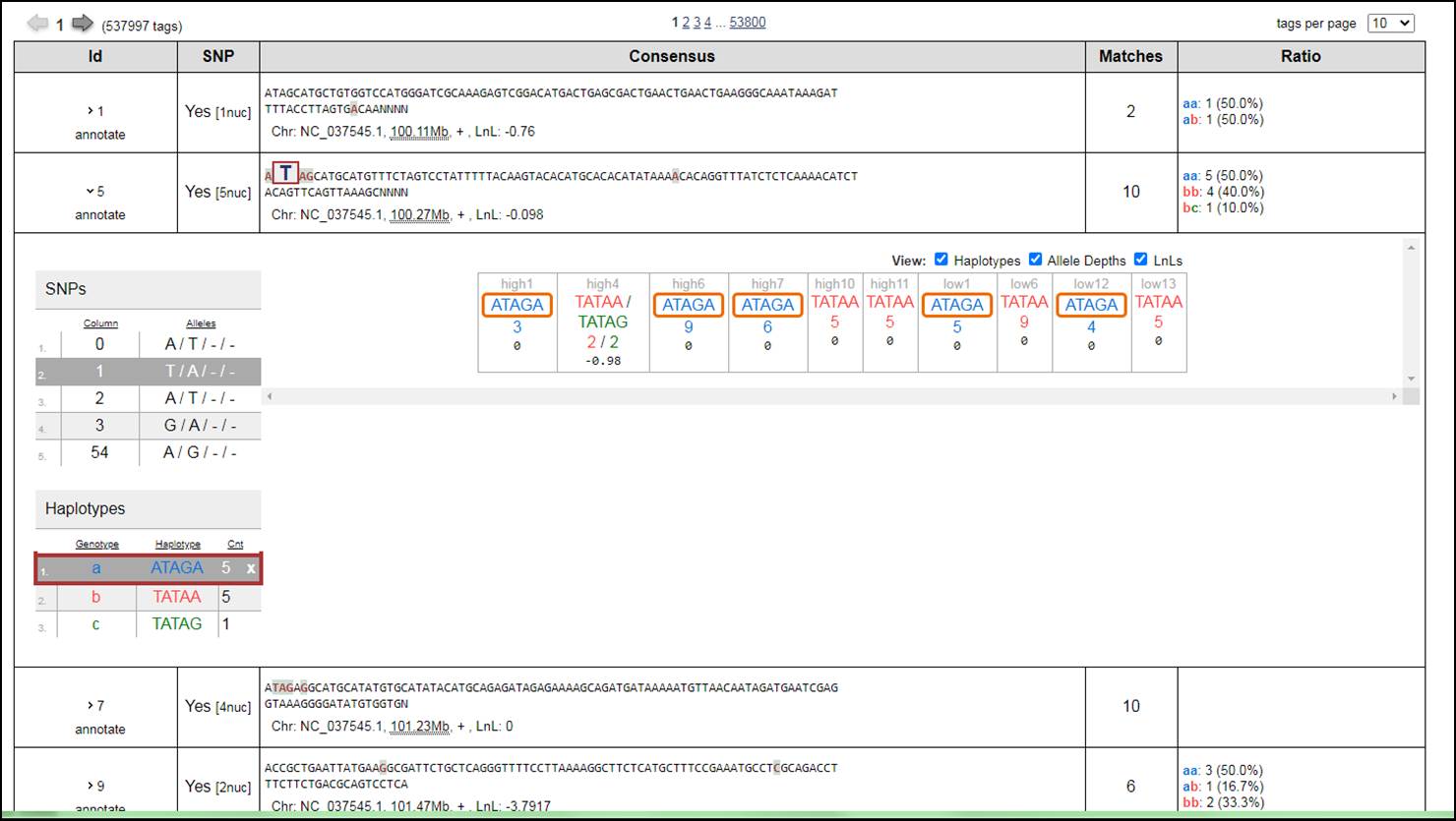
5. When click on any Catalog Id the information of the Summary Stats and Haplotype Stats are there.
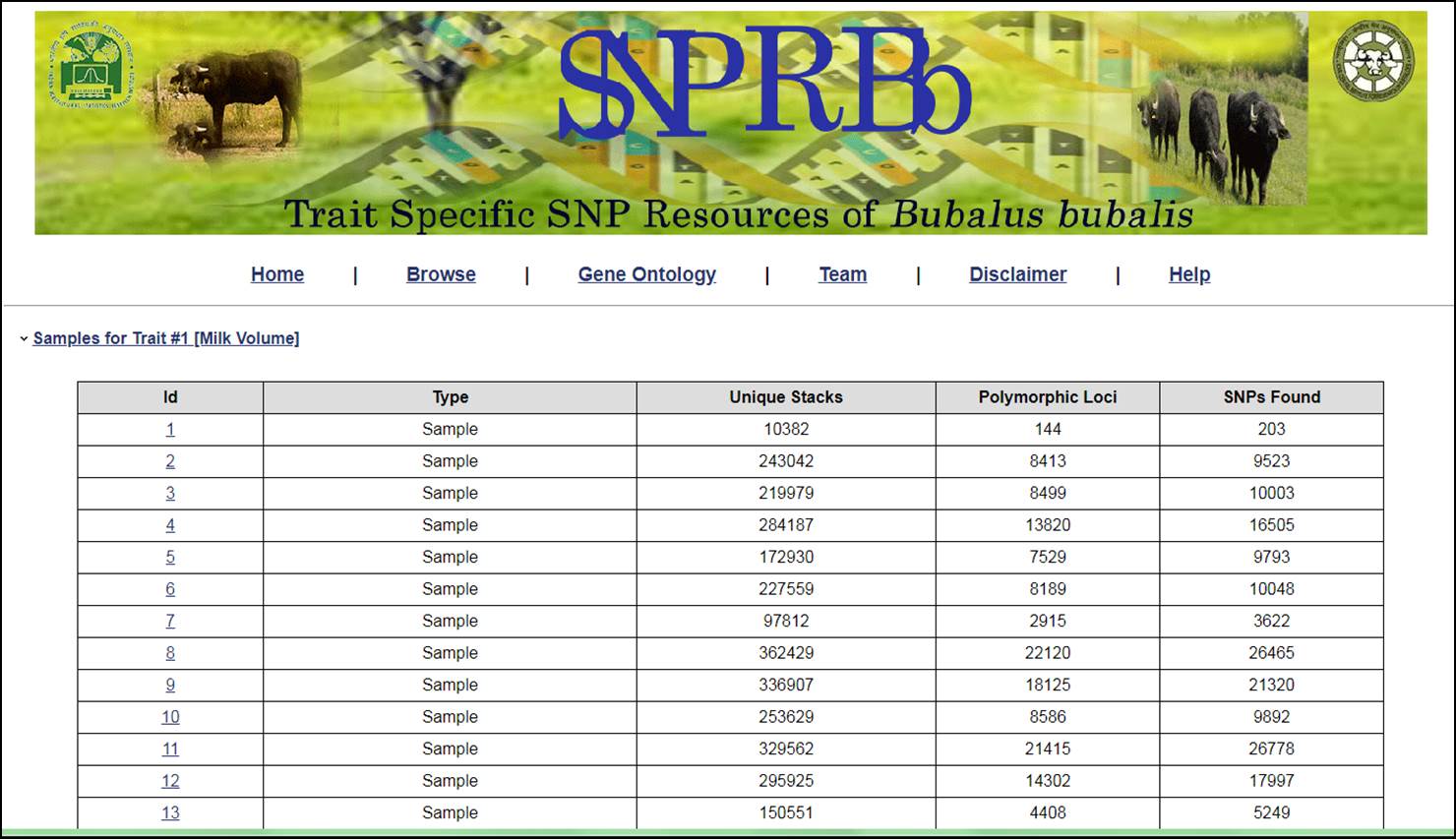
6. The identified SNPs can also be viewed as Sample wise by clicking on trait wise samples. This will display the information related to Sample Id as Sample, Unique Tags, Polymorphic Loci and Number of SNPs found in the sample.
7. Now the user can click on Sample id, details of the all the tags related to that sample will be displayed.
9. Filter by different parameters can be done.
10. We can execute both Id and catalog Id.
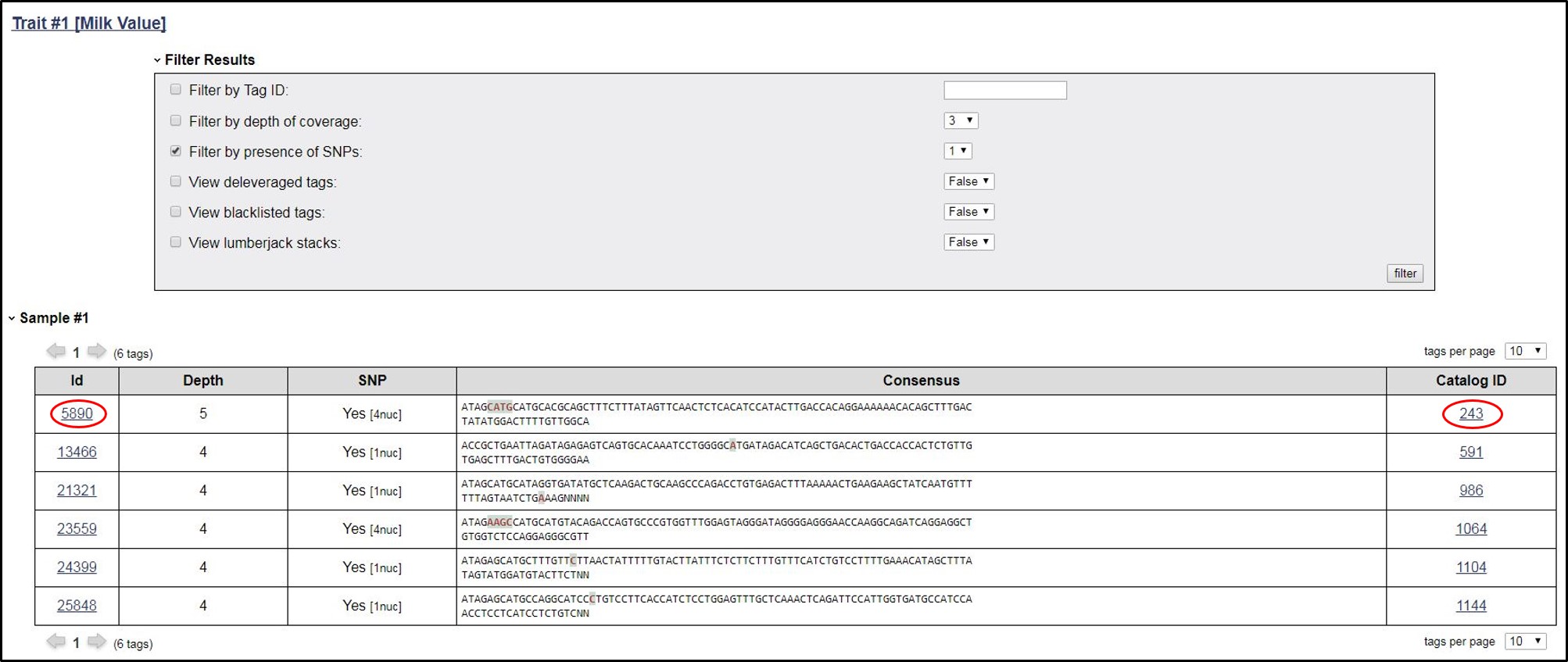
8. Now the user can click on Tag ID to get the other information that the belong to particular Catalog, Depth, Consensus model under Relationship including sequence ID and actual sequence alongwith the reference sequence of Bos taurus
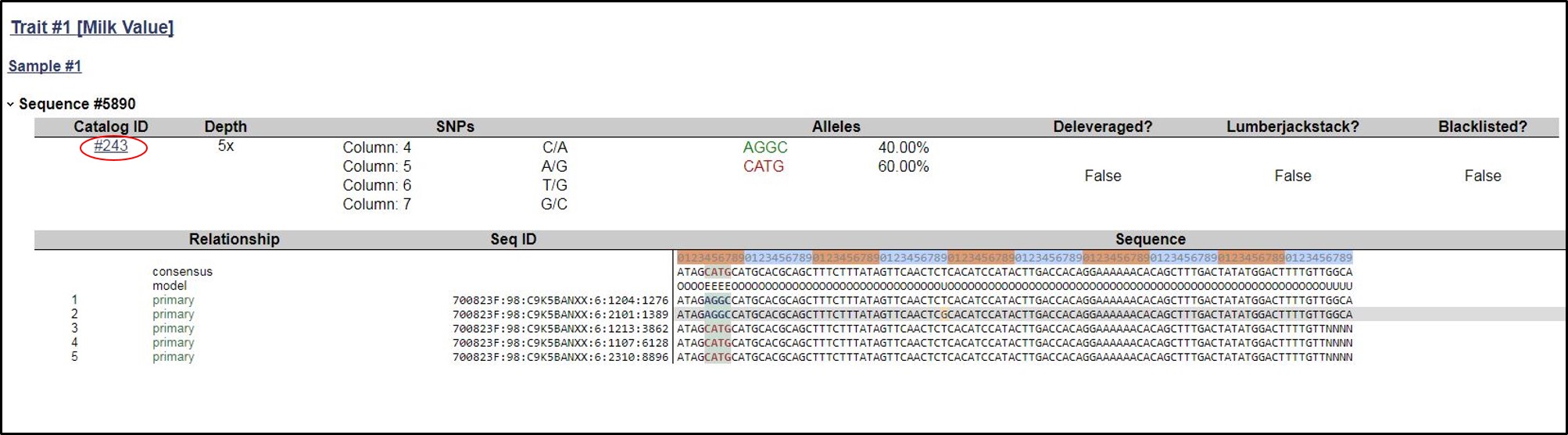
9. In this page, we can go back to the specified catalog by clicking on the Catalog Id.
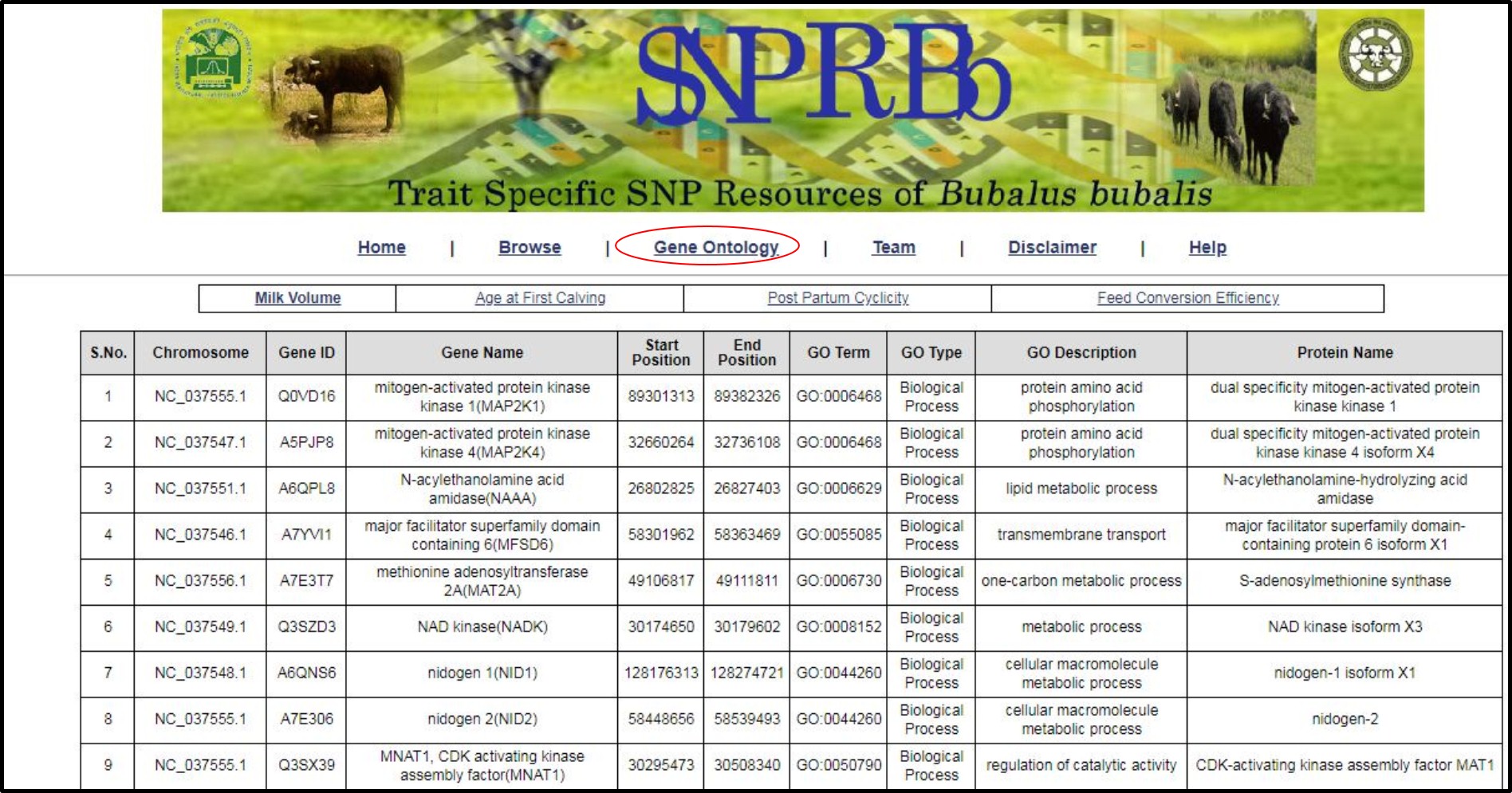
10. Gene ontology related information can be seen as mentioned in the following image.